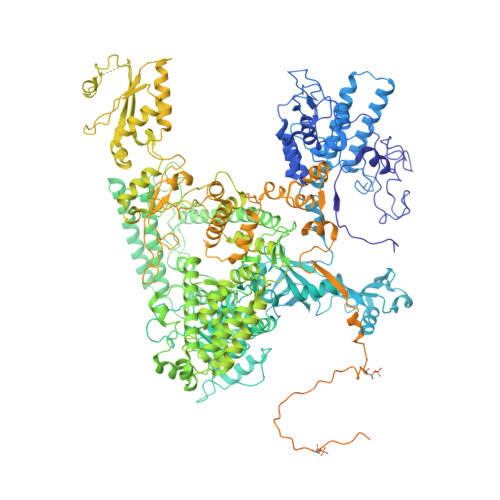
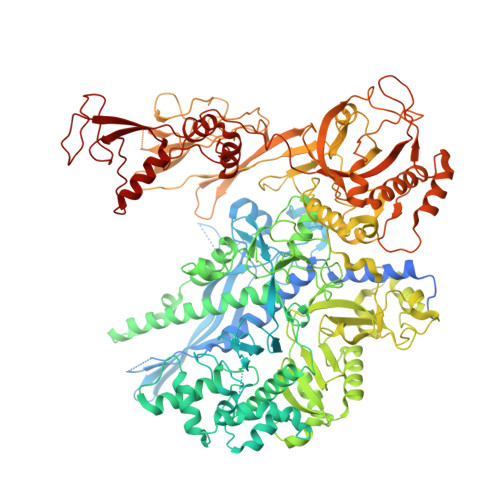
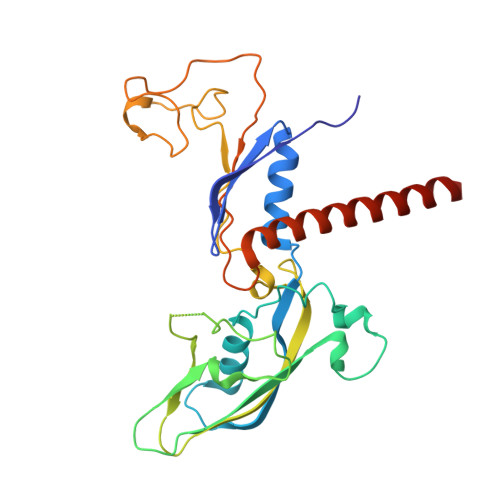
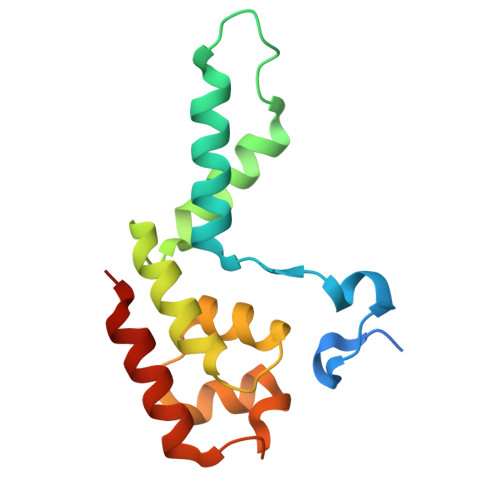
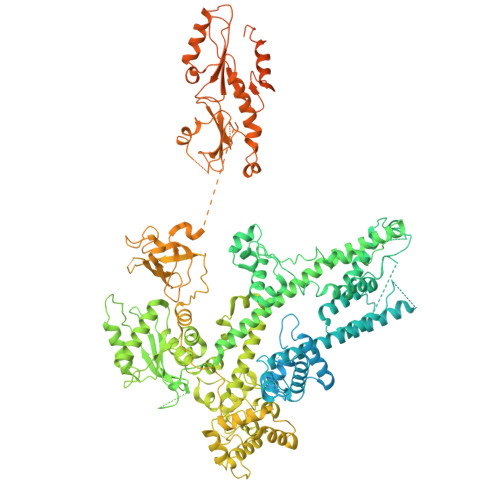
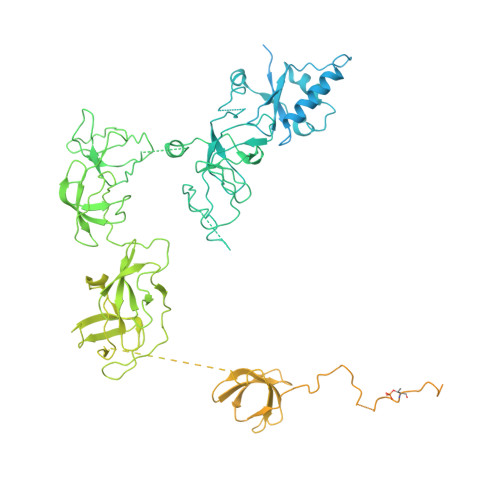
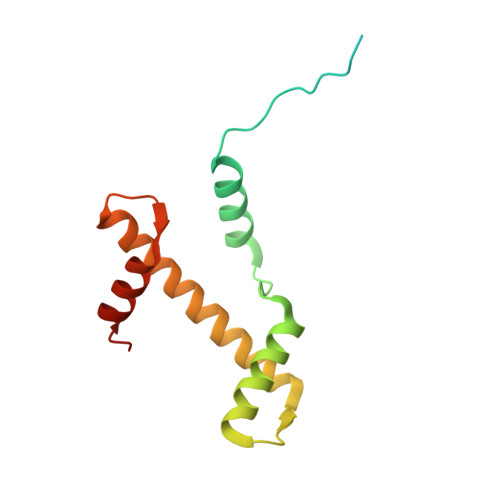
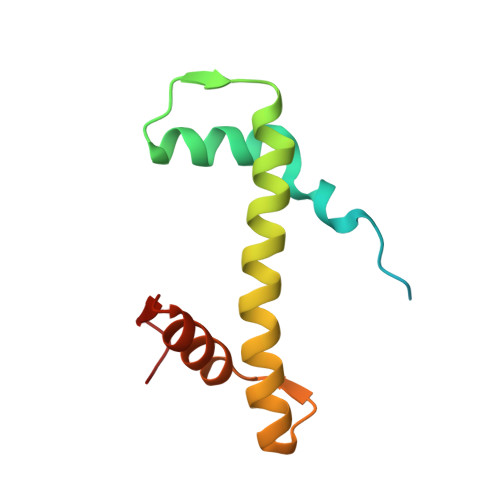
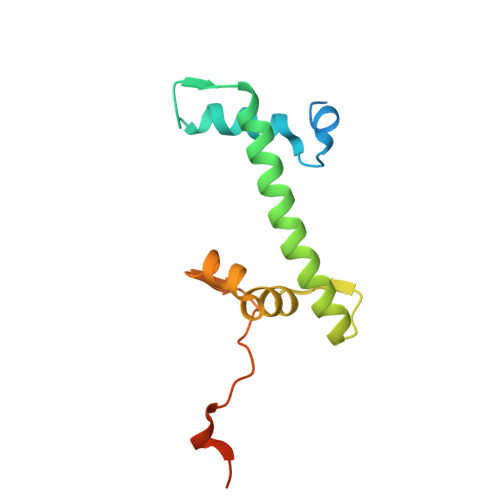
Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription.
Markert, J.W., Soffers, J.H., Farnung, L.(2024) Science : eadn6319-eadn6319
- PubMed: 39666822
- DOI: https://doi.org/10.1126/science.adn6319
- Primary Citation of Related Structures:
9EGX, 9EGY, 9EGZ, 9EH0, 9EH1, 9EH2 - PubMed Abstract:
During transcription, RNA polymerase II traverses through chromatin, and post-translational modifications including histone methylations mark regions of active transcription. Histone protein H3 lysine 36 trimethylation (H3K36me3), which is established by the histone methyltransferase SETD2, suppresses cryptic transcription, regulates splicing, and serves as a binding site for transcription elongation factors. The mechanism by which the transcription machinery coordinates the deposition of H3K36me3 is not well understood. Here we provide cryo-electron microscopy structures of mammalian RNA polymerase II-DSIF-SPT6-PAF1c-TFIIS-IWS1-SETD2-nucleosome elongation complexes, revealing that the transcription machinery regulates H3K36me3 deposition by SETD2 on downstream and upstream nucleosomes. SPT6 binds the exposed H2A-H2B dimer during transcription and the SPT6 death-like domain mediates an interaction with SETD2 bound to a nucleosome upstream of RNA polymerase II.
Organizational Affiliation:
Department of Cell Biology, Blavatnik Institute, Harvard Medical School, Boston, MA, USA.