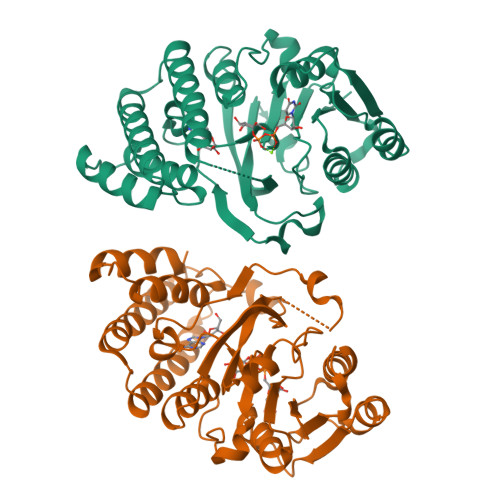
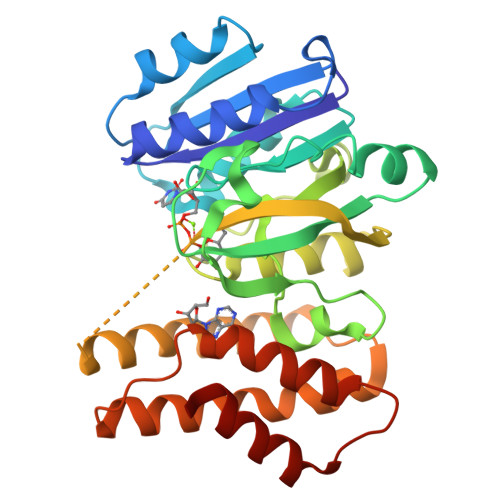
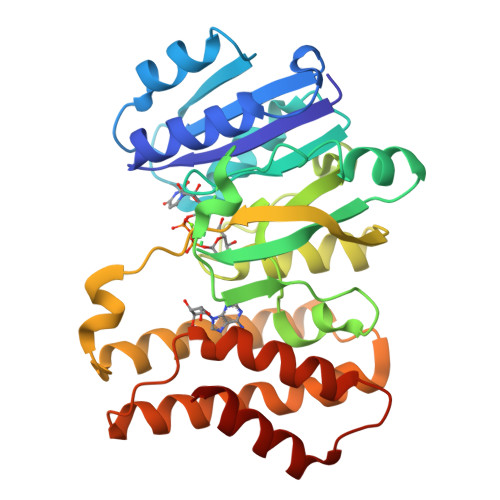
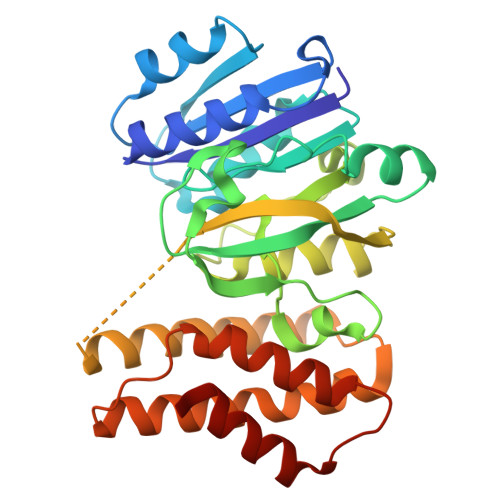
Efficient Synthesis of Glycodiversified Nucleoside Analogues by a Thermophilic Promiscuous Glycosyltransferase
Wang, Z., Li, J., Wang, X., Jin, B., Zhou, L., Zhao, Z., Gu, M., Song, X., Wang, J., Deng, Z., Wu, S., Zhang, Z., Chen, W.(2025) ACS Catal 15: 1217-1229