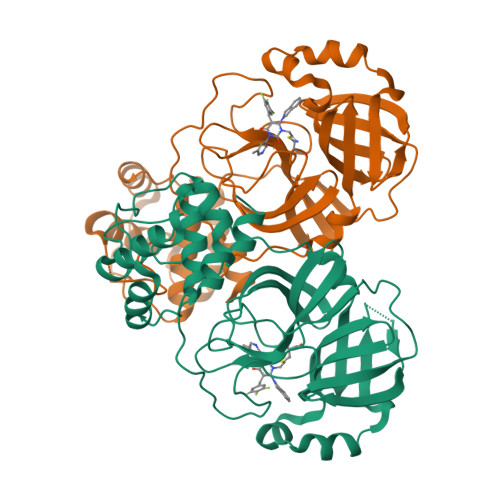
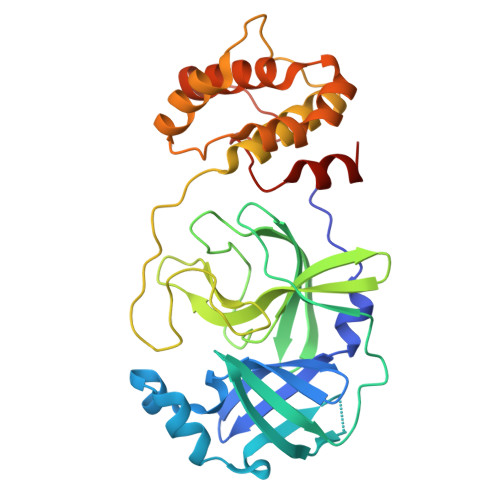
Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
Unoh, Y., Hirai, K., Uehara, S., Kawashima, S., Nobori, H., Sato, J., Shibayama, H., Hori, A., Nakahara, K., Kurahashi, K., Takamatsu, M., Yamamoto, S., Zhang, O., Tanimura, M., Dodo, R., Maruyama, Y., Sawa, H., Watari, R., Miyano, T., Kato, T., Sato, T., Tachibana, Y.To be published.