Fragment-Based Discovery of Novel Non-Hydroxamate LpxC Inhibitors with Antibacterial Activity.
Yamada, Y., Takashima, H., Walmsley, D.L., Ushiyama, F., Matsuda, Y., Kanazawa, H., Yamaguchi-Sasaki, T., Tanaka-Yamamoto, N., Yamagishi, J., Kurimoto-Tsuruta, R., Ogata, Y., Ohtake, N., Angove, H., Baker, L., Harris, R., Macias, A., Robertson, A., Surgenor, A., Watanabe, H., Nakano, K., Mima, M., Iwamoto, K., Okada, A., Takata, I., Hitaka, K., Tanaka, A., Fujita, K., Sugiyama, H., Hubbard, R.E.(2020) J Med Chem 63: 14805-14820
- PubMed: 33210531
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01215
- Primary Citation of Related Structures:
7CI4, 7CI5, 7CI6, 7CI7, 7CI8, 7CI9, 7CIA, 7CIB, 7CIC, 7CID, 7CIE - PubMed Abstract:
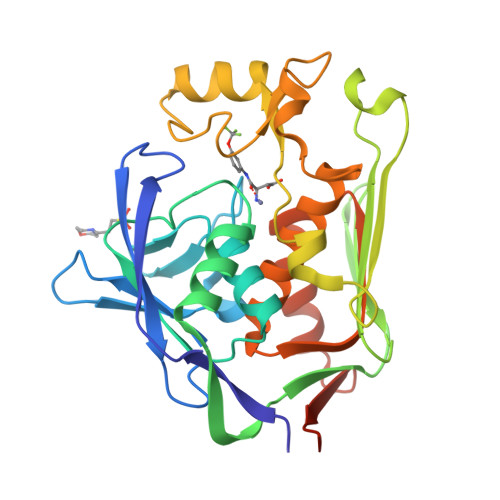
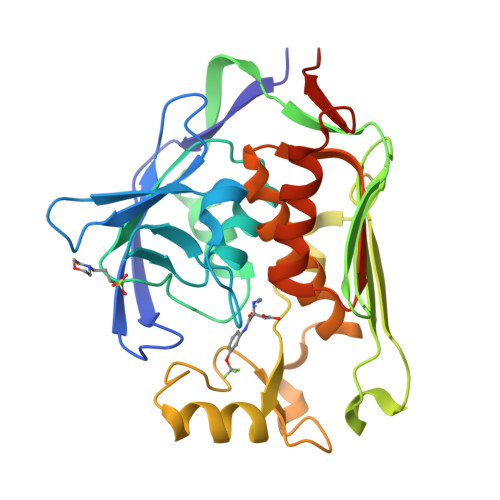
UDP-3- O -acyl- N -acetylglucosamine deacetylase (LpxC) is a zinc metalloenzyme that catalyzes the first committed step in the biosynthesis of Lipid A, an essential component of the cell envelope of Gram-negative bacteria. The most advanced, disclosed LpxC inhibitors showing antibacterial activity coordinate zinc through a hydroxamate moiety with concerns about binding to other metalloenzymes. Here, we describe the discovery, optimization, and efficacy of two series of compounds derived from fragments with differing modes of zinc chelation. A series was evolved from a fragment where a glycine moiety complexes zinc, which achieved low nanomolar potency in an enzyme functional assay but poor antibacterial activity on cell cultures. A second series was based on a fragment that chelated zinc through an imidazole moiety. Structure-guided design led to a 2-(1 S -hydroxyethyl)-imidazole derivative exhibiting low nanomolar inhibition of LpxC and a minimum inhibitory concentration (MIC) of 4 μg/mL against Pseudomonas aeruginosa , which is little affected by the presence of albumin.
Organizational Affiliation:
Taisho Pharmaceutical Co., Ltd., Saitama 331-9530, Japan.