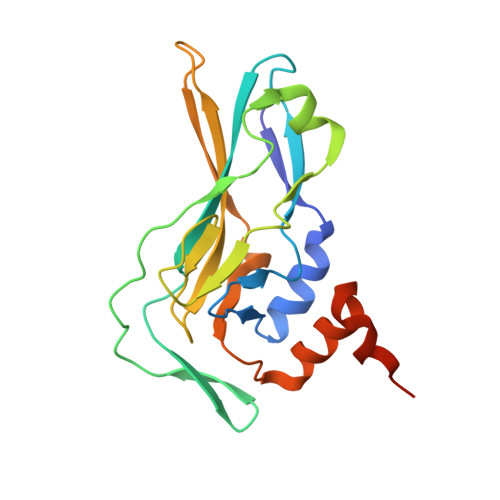
Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Newman, J.A., Gavard, A.E., Imprachim, N., Aitkenhead, H., Sheppard, H.E., Te Poele, R., Clarke, P.A., Hossain, M.A., Temme, L., Oh, H.J., Wells, C.I., Davis-Gilbert, Z.W., Workman, P., Gileadi, O., Drewry, D.H.(2025) Nat Commun 16: 1596-1596
- PubMed: 39952925
- DOI: https://doi.org/10.1038/s41467-025-56213-1
- Primary Citation of Related Structures:
6F58, 6F59, 7ZK2, 7ZKF, 7ZL2, 8A10, 8A7N, 8CDN - PubMed Abstract:
Brachyury is a transcription factor that plays an essential role in tumour growth of the rare bone cancer chordoma and is implicated in other solid tumours. Brachyury is minimally expressed in healthy tissues, making it a potential therapeutic target. Unfortunately, as a ligandless transcription factor, brachyury has historically been considered undruggable. To investigate direct targeting of brachyury by small molecules, we determine the structure of human brachyury both alone and in complex with DNA. The structures provide insights into DNA binding and the context of the chordoma associated G177D variant. We use crystallographic fragment screening to identify hotspots on numerous pockets on the brachyury surface. Finally, we perform follow-up chemistry on fragment hits and describe the progression of a thiazole chemical series into binders with low µM potency. Thus we show that brachyury is ligandable and provide an example of how crystallographic fragment screening may be used to target protein classes that are difficult to address using other approaches.
Organizational Affiliation:
Centre for Medicines Discovery, University of Oxford, Oxford, UK. Joseph.newman@cmd.ox.ac.uk.