Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies.
Harshbarger, W.D., Holzapfel, G., Seraj, N., Tian, S., Chesterman, C., Fu, Z., Pan, Y., Harelson, C., Peng, D., Huang, Y., Chandramouli, S., Malito, E., Bottomley, M.J., Williams, J.(2024) Vaccines (Basel) 12
- PubMed: 39460278
- DOI: https://doi.org/10.3390/vaccines12101111
- Primary Citation of Related Structures:
8V5L, 8V5P, 8V5Q, 8V5S - PubMed Abstract:
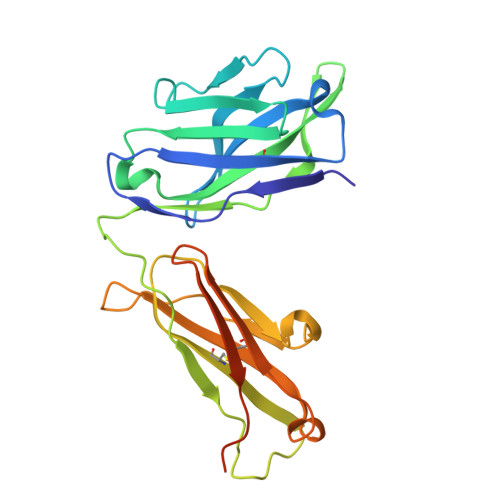
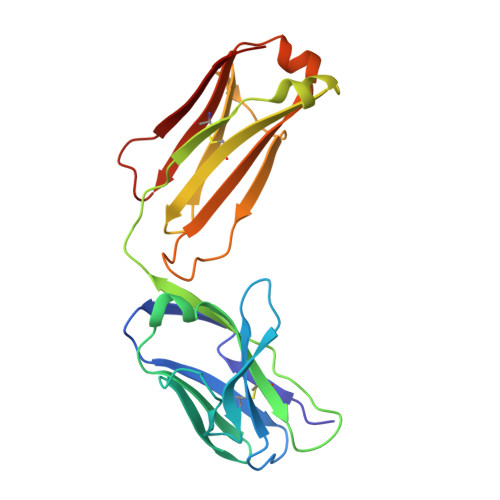
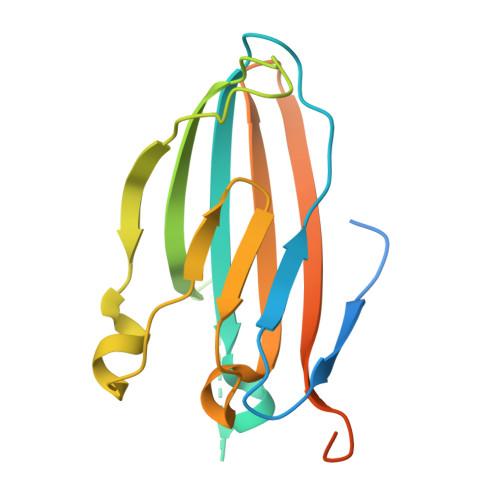
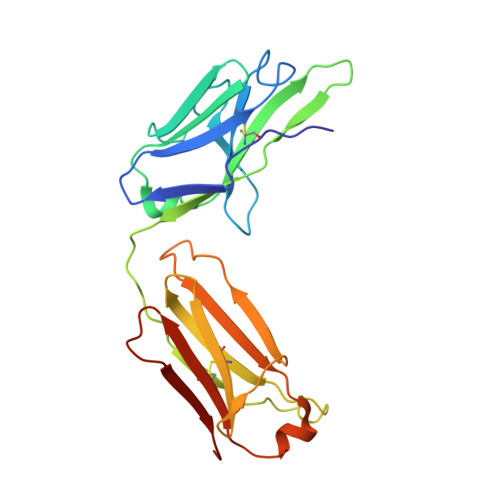
Background: Varicella zoster virus (VZV) is the causative agent for chickenpox and herpes zoster (HZ, shingles). HZ is a debilitating disease affecting elderly and immunocompromised populations. Glycoprotein E (gE) is indispensable for viral replication and cell-to-cell spread and is the primary target for anti-VZV antibodies. Importantly, gE is the sole antigen in Shingrix, a highly efficacious, AS01 B -adjuvanted vaccine approved in multiple countries for the prevention of HZ, yet the three-dimensional (3D) structure of gE remains elusive. Objectives : We sought to determine the structure of VZV gE and to understand in detail its interactions with neutralizing antibodies. Methods : We used X-ray crystallography and cryo-electron microscopy to elucidate structures of gE bound by recombinant Fabs of antibodies previously elicited through vaccination with Zostavax, a live, attenuated vaccine. Results : The 3D structures resolve distinct central and C-terminal antigenic domains, presenting an array of diverse conformational epitopes. The central domain has two beta-sheets and two alpha helices, including an IgG-like fold. The C-terminal domain exhibits 3 beta-sheets and an Ig-like fold and high structural similarity to HSV1 gE. Conclusions : gE from VZV-infected cells elicits a human antibody response with a preference for the gI binding domain of gE. These results yield insights to VZV gE structure and immunogenicity, provide a framework for future studies, and may guide the design of additional herpesvirus vaccine antigens. Teaser: Structures of varicella zoster virus glycoprotein E reveal distinct antigenic domains and define epitopes for vaccine-elicited human antibodies.
Organizational Affiliation:
GSK, Rockville, MD 20850, USA.