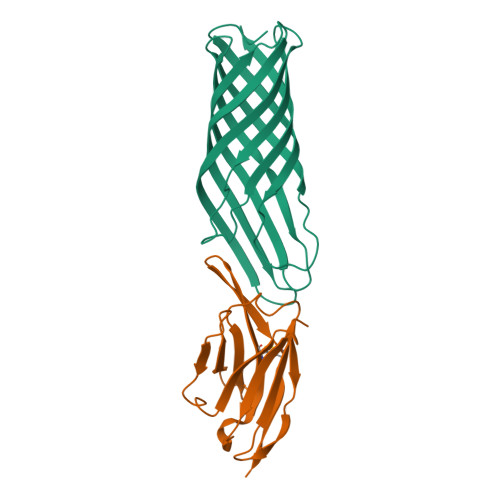
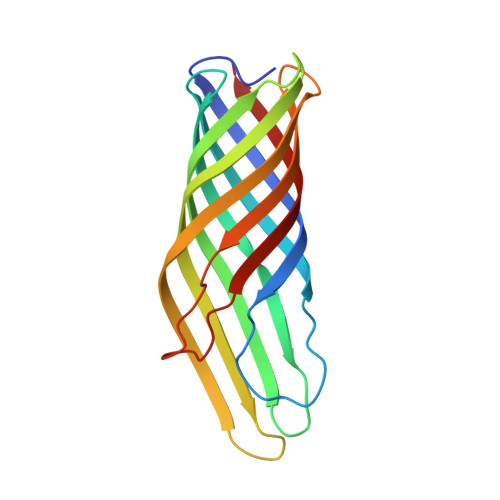
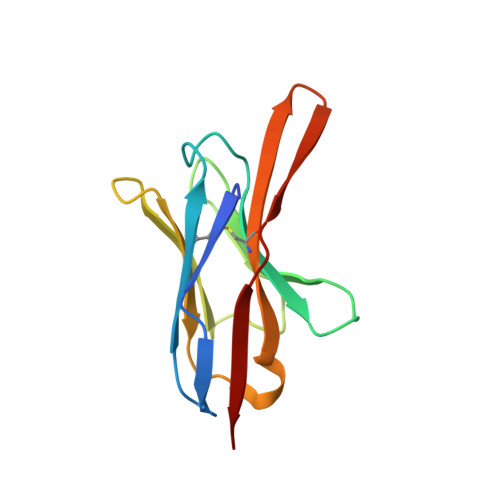
Rapid detection and capture of clinical Escherichia coli strains mediated by OmpA-targeting nanobodies.
Sorgenfrei, M., Hurlimann, L.M., Printz, A., Wegner, F., Morger, D., Ackle, F., Remy, M.M., Montowski, G., Keserue, H.A., Cuenod, A., Imkamp, F., Egli, A., Keller, P.M., Seeger, M.A.(2025) Commun Biol 8: 1047-1047
- PubMed: 40659774
- DOI: https://doi.org/10.1038/s42003-025-08345-9
- Primary Citation of Related Structures:
9FZC, 9FZD - PubMed Abstract:
Escherichia coli is a major cause of blood stream and urinary tract infections. Owing to the spread of antimicrobial resistance, it is often treated with an inadequate antibiotic. With the aim to accelerate the diagnostics of this key pathogen, we used the flycode technology to generate nanobodies against the conserved and highly abundant outer membrane protein OmpA. Two nanobodies each recognizing a different isoform of OmpA were shown by flow cytometry to recognize > 91% of 85,680 E. coli OmpA sequences deposited in a large bacterial genome database. Crystal structures of these nanobodies in complex with the respective OmpA isoform revealed interactions with all four surface accessible loops of OmpA. Steric hindrance caused by dense O-antigen layers initially impeded reliable capture of clinical E. coli strains. By generating nanobody constructs with long linkers and by thinning the O-antigen layer through alterations to growth medium and buffers, we achieved to capture < 50 CFU/mL. Our work provides a framework to generate nanobodies for the specific and sensitive detection and capture of clinically relevant pathogenic bacteria.
- Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.
Organizational Affiliation: