Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification.
Cho, G., Lee, J., Kim, J.(2023) Nucleic Acids Res 51: 1971-1983
- PubMed: 36762482
- DOI: https://doi.org/10.1093/nar/gkad048
- Primary Citation of Related Structures:
8H0S, 8H0T, 8H1A, 8H1B, 8H27 - PubMed Abstract:
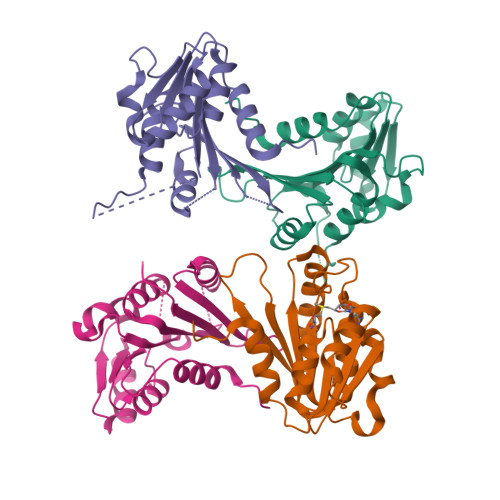
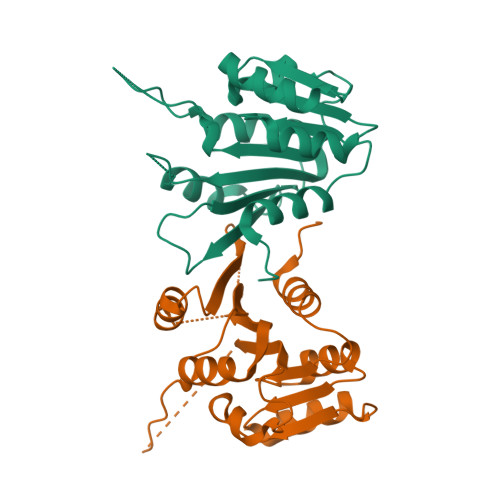
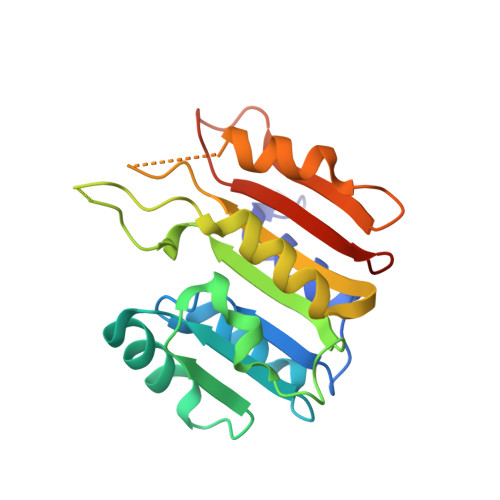
The uridine at the 34th position of tRNA, which is able to base pair with the 3'-end codon on mRNA, is usually modified to influence many aspects of decoding properties during translation. Derivatives of 5-methyluridine (xm5U), which include methylaminomethyl (mnm-) or carboxymethylaminomethyl (cmnm-) groups at C5 of uracil base, are widely conserved at the 34th position of many prokaryotic tRNAs. In Gram-negative bacteria such as Escherichia coli, a bifunctional MnmC is involved in the last two reactions of the biosynthesis of mnm5(s2)U, in which the enzyme first converts cmnm5(s2)U to 5-aminomethyl-(2-thio)uridine (nm5(s2)U) and subsequently installs the methyl group to complete the formation of mnm5(s2)U. Although mnm5s2U has been identified in tRNAs of Gram-positive bacteria and plants as well, their genomes do not contain an mnmC ortholog and the gene(s) responsible for this modification is unknown. We discovered that MnmM, previously known as YtqB, is the methyltransferase that converts nm5s2U to mnm5s2U in Bacillus subtilis through comparative genomics, gene complementation experiments, and in vitro assays. Furthermore, we determined X-ray crystal structures of MnmM complexed with anticodon stem loop of tRNAGln. The structures provide the molecular basis underlying the importance of U33-nm5s2U34-U35 as the key determinant for the specificity of MnmM.
Organizational Affiliation:
Department of Chemistry, Gwangju Institute of Science and Technology, Gwangju 61005, Korea.