News
Rapid Visualization of Large Structure using NGL/MMTF
08/23
Fast, interactive display of molecular complexes containing millions of atoms is now possible, without plug-ins, on desktop computers and smartphones. The new NGL Viewer enables easy visualization of even the largest structures for everyone in research and education.
In addition, the new binary compressed Macromolecular Transmission Format (MMTF.rcsb.org) massively reduces network transfer and parsing times. With these advances, we are prepared for upcoming advances in experimental techniques that are already delivering structures of rapidly increasing size and complexity.
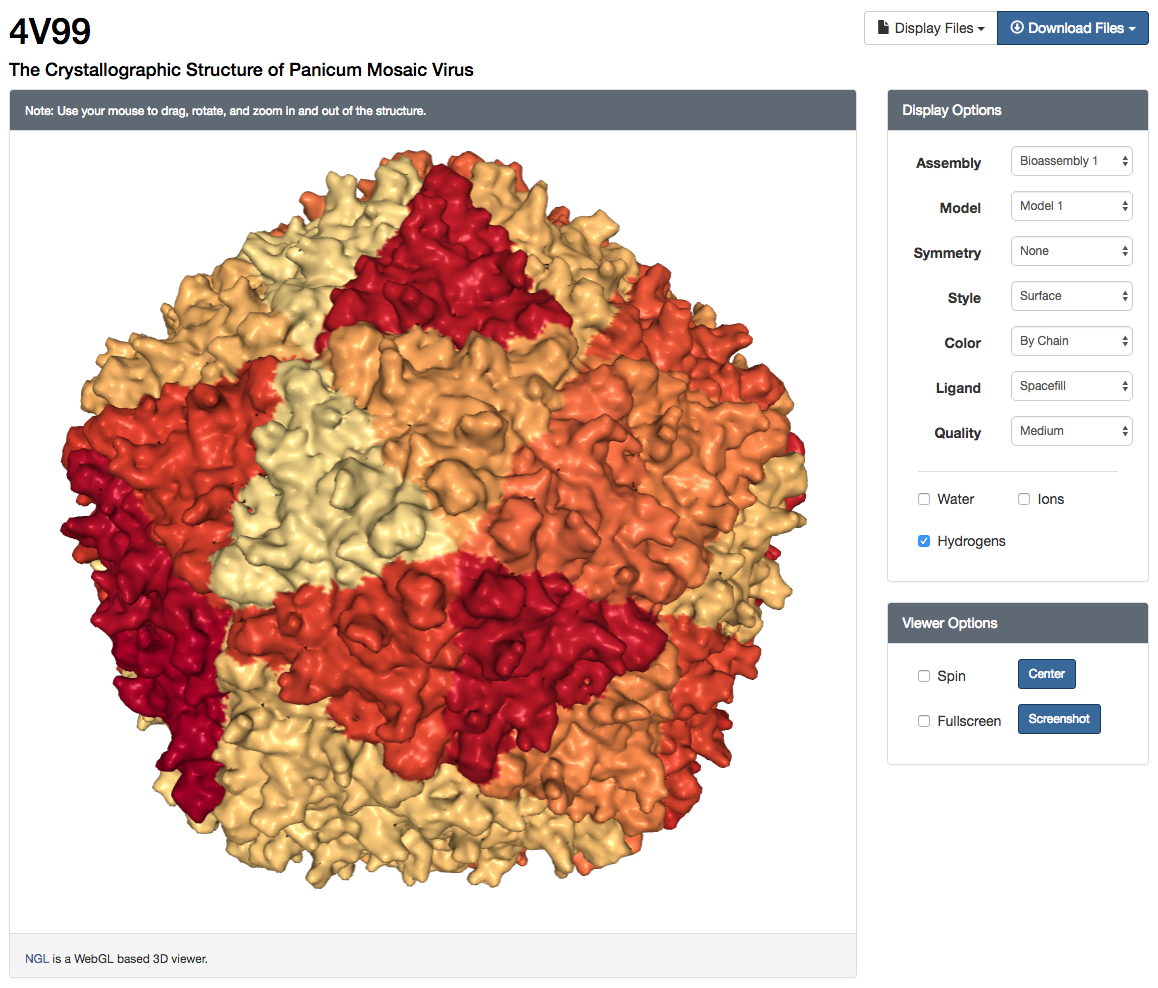 NGL Viewer user interface and surface rendering of "The Crystallographic Structure of Panicum Mosaic Virus" (4V99)
NGL Viewer user interface and surface rendering of "The Crystallographic Structure of Panicum Mosaic Virus" (4V99)Launching & Using the NGL Viewer
The NGL Viewer can be launched from the 3D view tab or under the image display box on each Structure Summary page (e.g., PDB ID 4HHB). NGL is the only supported viewer for large structures comprising more than 10,000 residues in either the asymmetric unit or the biological assemblyl (e.g., PDB ID 4V99). For structures with a large asymmetric unit, a backbone-only structure is displayed by default to improve loading time. Display style and quality are optimized so the viewer will run smoothly on both mobile and desktop systems, but can be customized for more powerful systems or for the generation of high resolution images.
NGL Features
- Viewer options (Spin, Center, Fullscreen, Screenshot)
- Basic symmetry support (Local & global symmetry axes)
- Options for Protein/RNA/DNA Display (Backbone, Surface, Cartoon, Spacefill, Licorice)
- Options for Ligand Display (Ball+Stick, Spacefill)
- Multiple color schemes (By Chain, Rainbow (default), Element, B-Factor, Secondary Structure, Hydrophobicity)
- Rendering Quality Options Automatic, Low, Medium, High)
- Optional Display (Water, Hydrogens, Ions)
- Assembly support (Asymmetric Unit, Biological Assemblies, Crystallographic Cells)
- Model support (NMR & multi-model X-ray structures)
Crystallographic Cells
Crystallographic unit cells can be displayed, as well as “supercells” comprising a set of adjacent unit cells for X-ray entries.
.png) Unit cell representation of the "Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin" (PDB ID 3PQR)
Unit cell representation of the "Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin" (PDB ID 3PQR).png) Supercell representation of the "Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin" (3PQR)
Supercell representation of the "Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin" (3PQR)Reference Links
- Macromolecular Transmission Format (MMTF) - http://mmtf.rcsb.org/
- Web-based molecular graphics for large complexes. AS Rose, AR Bradley, Y Valasatava, JM Duarte, A Prlic and PW Rose. 2016. In Proceedings of the 21st International Conference on Web3D Technology (Web3D '16). ACM, New York, NY, USA, 185-186. DOI: 10.1145/2945292.2945324












